Neighborhood Explorer Documentation
|
Neighborhood Explorer: initial appearance
|
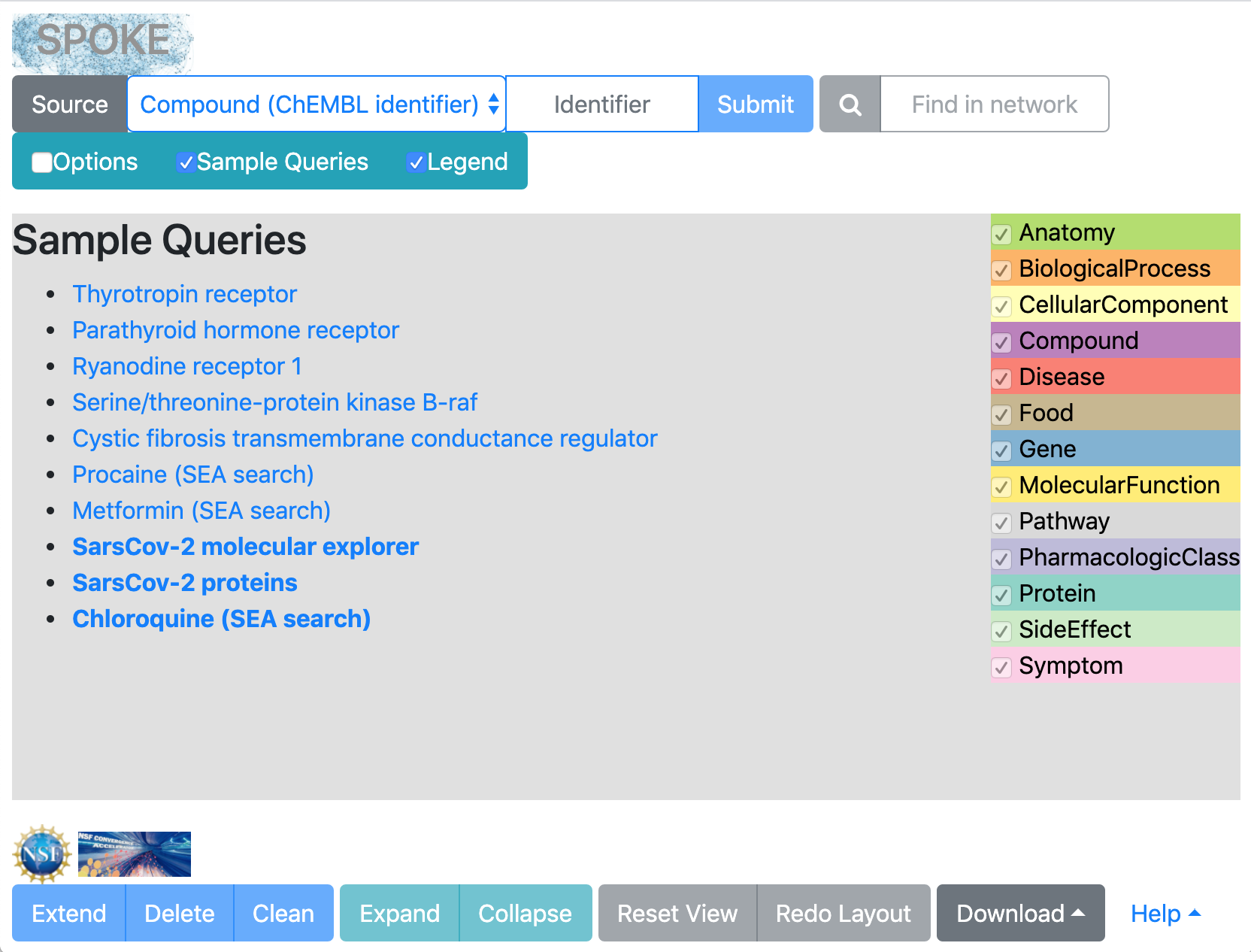 |
[back to top]
Introduction
SPOKE
(Scalable Precision Medicine Knowledge Engine)
is a very large network containing multiple types of biological data.
Pooling such diverse data into a single knowledge environment
allows identifying new connections, with implications for
biomedical applications like personalized medicine:
suggesting which drugs may be effective for a specific patient.
An earlier version of the network was used to suggest new uses for existing drugs
(Himmelstein
et al., eLife 2017 Sep 22;6. pii: e26726).
SPOKE is a heterogeneous network,
meaning that different nodes (points) within the network
can represent different types of data.
The edges between pairs of nodes represent known connections.
Paths that follow a series of edges may connect nodes not
previously known to be related.
SPOKE is much too large and dense to comprehend visually all at once.
Although automated analyses may operate on the entire network, a graphical
interface for exploring limited areas or subsets is need for human interaction.
The Neighborhood Explorer allows interactively
finding and viewing specific “neighborhoods” within SPOKE.
As detailed below, it allows searching SPOKE for a specific drug compound,
gene, or protein, and seeing what other nodes are in its immediate neighborhood,
connected by one or a few edges. These other nodes could be any of the
data types in the heterogeneous network, including related diseases,
side effects, pathways, and other compounds, genes, or proteins.
The search can be restricted
to certain node and edge types, and by edge value.
The resulting network is displayed with color-coding by node type,
with detailed information for any node or edge popping up on mouseover.
Sets of terminal nodes (“leaf” nodes) are grouped into
rectangles that can be collapsed to simplify the view.
The network can be extended further from any
node(s) of interest by one or more additional rounds of searching.
[back to top]
Searching and Search Options
All search fields except for SEA searches will accept any text that matches
the name, or identifier of the item being searched. Once a full text query is entered,
the matching values will be displayed in a pop-down windows. Selecting one of the values
will place the required identifier or name in the search box.
Search types, with the search type in parentheses:
- Compound (ChEMBL identifier)
– a drug or other chemical compound specified by its
identifier in ChEMBL,
for example: CHEMBL657 for diphenhydramine
- Disease (DO identifier)
– a disease or condition specified by its identifier in
Disease Ontology,
for example: DOID:3650 for lactic acidosis.
Full text search is generally an easier approach.
- EC (Enzyme Commission identifier)
– an EC number, for example 4.1.3.3 for N-acetylneuraminate lyase
- Food (Food identifier)
– A Food Ontology (https://foodon.org) term. For example,
FOODON:00001244 for "coffee beverage". Full text search is generally an easier approach.
- Gene (Entrez name)
– a human gene specified by its name in
Entrez Gene,
for example: DMD for human dystrophin
- Node type – all nodes of the specified type;
generally not feasible except for SARSCov2
(SARS-CoV-2 proteins, a relatively small set)
- Organism (Species name)
– for example, Lactobacillus casei
- Pathway (Pathway name)
– for example, L-tryptophan degradation IX
- Protein (UniProt identifier)
– a protein specified by its identifier in
UniProtKB,
for example: P28223 for the human 5-hydroxytryptamine receptor 2A
- Protein (UniProt name)
– a protein specified by its name in
UniProtKB,
for example: 5HT2A_HUMAN for the human 5-hydroxytryptamine receptor 2A
- Reaction (Reaction identifier)
– a reaction identifier from KEGG or MetaCyc.
Full text search is generally an easier approach.
- SEA (SMILES)
– a compound specified by
SMILES string for a two-stage search:
- Similarity
Ensemble Approach search to predict proteins that might
bind the compound (see Keiser et al.,
Nat Biotechnol 25:197 (2007))
- SPOKE neighborhood search with all of the resulting proteins as queries,
as well as the initial compound
Doing a SEA search with path-length set to zero in the
options gives only the edges from the first stage
described above.
The identifier to enter is rarely known in advance.
Clicking the Source button shows the website corresponding to the
current search type, for example,
Disease Ontology
for diseases. The identifier(s) can be looked up at this website.
For SMILES string lookup, however, sites other than the
SEA website
are recommended, such as PubChem Compound or
ChEMBL.
SMILES string example, for
lurasidone:
C1CCC(C(C1)CN2CCN(CC2)C3=NSC4=CC=CC=C43)CN5C(=O)C6C7CCC(C7)C6C5=O
|
results from CFTR sample query (Apr 2020)
|
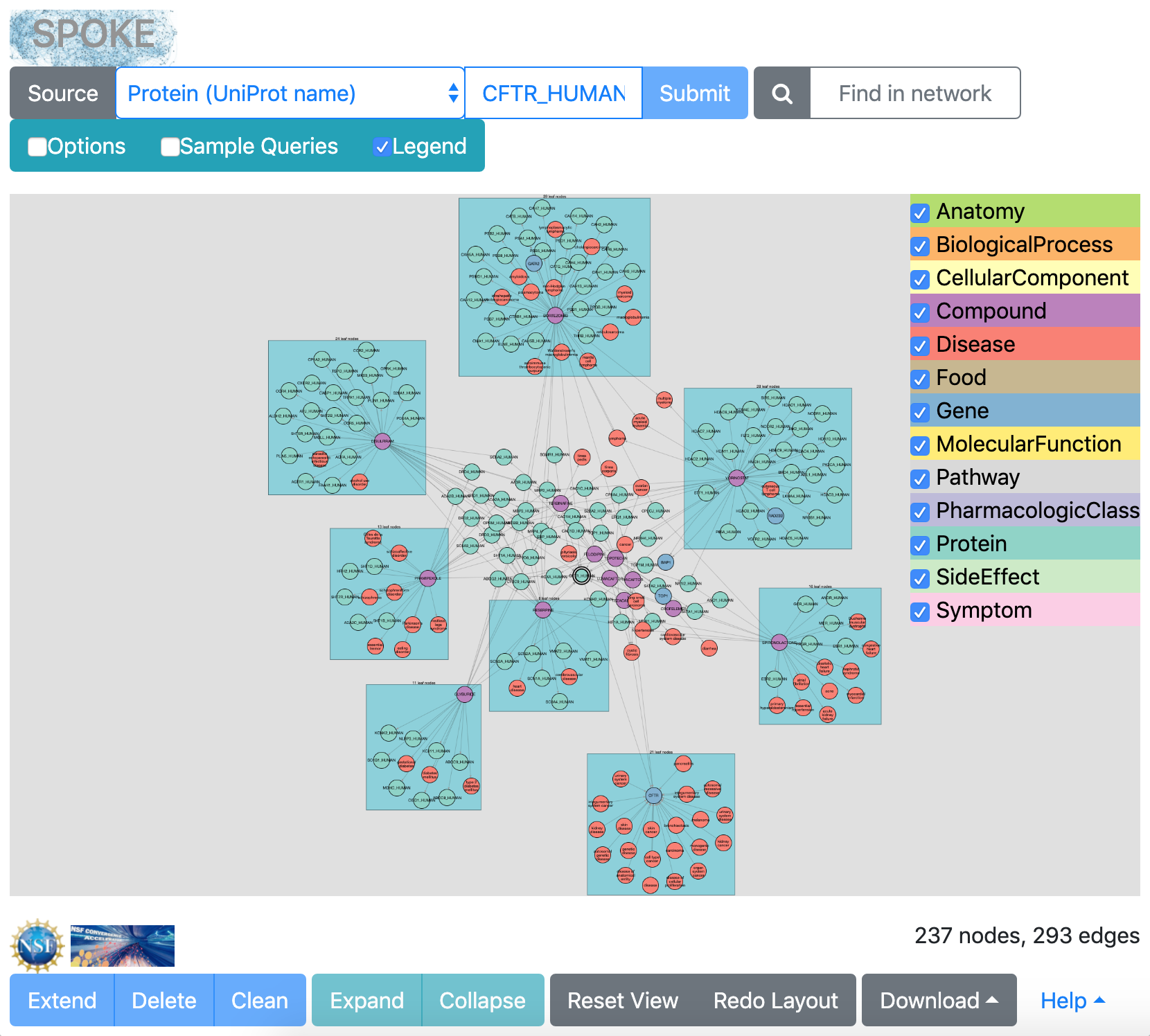
|
Entering the exact match to long data type such as a pathway or species name
is often very difficult, and sometimes a broader search is desired.
For these reasons, in addition to the full-text search mentioned above,
all search types except SEA and Node type
allow entering a partial match as a
regular expression.
A leading tilde symbol ~ is required to indicate
when a regular expression is being used. Examples:
- ~.*fatty acid.* in a pathway search to find all pathways with
name containing “fatty acid”
- ~Akkermansia.* in an organism search to find all taxa with name
starting with “Akkermansia”
**It is very important to set options
before submitting the search,
mainly to limit the results to a reasonable number of nodes and edges.
Searches that are too broad not only take longer,
the results may be impossible to view.
A good way to get a feel for a reasonable amount of results and the
corresponding option settings is to run some of the
sample queries.
Checkboxes show/hide parts of the interface:
-
Options – filters and other search settings:
- Node and Edge Types
- Show nodes
[several checkboxes controlling which node types to include in the results]
- Show edges
[several checkboxes controlling which edge types to include in the results]
- Node and Edge Attributes
- Filter nodes
- Compound max phase >=[minPhase]
– minimum value of the maximum clinical trial phase a compound has reached
(for treating any disease); for example, 4 to exclude compounds that have
not been approved as treatments. Max phase is part of the compound data from
ChEMBL.
- BV-BRC Organisms with AMR Phenotype: Check this box to include only
BV-BRC
organisms with antimicrobial resistance (AMR) phenotypes.
- BV-BRC Organisms where host is human: Check this box to include only
BV-BRC
organisms isolated from human hosts.
- BV-BRC Organisms exhibiting Resistance Phenotype: Check this box to include only
BV-BRC
organisms exhibiting a resistance phenotype.
- BV-BRC Organisms isolated in the US: Check this box to include only
BV-BRC
organisms isolated in the US.
- Protein is an enzyme: Check this box to only include proteins that are enzymes.
- Protein is virulence factor: Check this box to only include proteins classified
as virulence factors according
to VFDB.
- UniProt Protein source: TrEMBL (uncurated) and/or
SwissProt (curated)
- Filter edges
- AnatomyCellType-expresses-Gene expression level
– include edges with expression not detected, low,
medium, and/or high according to the
Human Protein Atlas
- Compound-treats-Disease phase >=[minPhase]
– minimum value of the maximum clinical trial phase the compound
has reached as a treatment for the specific disease
- Disease-associates-Gene DISEASES sources
– type of evidence from the
DISEASES
database: textmining, knowledge, and/or experiments
- Compound-Clinical_Trial-Disease CTKP purpose
– main purpose of the clinical trial. This information comes from
AACT
This filter is applied to the following edge types: Compound-mentioned_clinical_trials_for-Disease, Compound-in_clinical_trials_for-Disease and Compound-treats-Disease
- Compound-UPREGULATES/DOWNREGULATES-Gene DOSE_LINCS1000 times
– Time point used for the calculation of efficacy and potency. This information comes from
DOSE-L1000
This filter is applied to the following edge types: Compound-upregulates-Gene and Compound-downregulates-Gene
- Compound-UPREGULATES/DOWNREGULATES-Gene DOSE_LINCS1000 tissues
– The tissue to which the cell line used for the calculation of efficacy and potency belongs. The default option is a Summary edge, which contains for that compound-gene pair, all the tissues and time points tested. This information comes from
DOSE-L1000
This filter is applied to the following edge types: Compound-upregulates-Gene and Compound-downregulates-Gene
- Disease-associates-Gene text mining z-score >=[minZ]
- Protein-interacts-Protein score >=[minScore]
- Limits
- Maximum path length <=[L]
– number of edges outward from the query node to include in the results;
1 is usually recommended, 2 if including only a few node and edge types.
The result network can be extended later.
- SEA p-Value Cutoff <=1e-[N]
– significance of protein hits from SEA search
- Graphics limit (nodes + edges) [K]
– limit at which to warn the user of too many results to render
(default 2000)
-
Sample Queries – a series of sample queries, any of which
can be clicked to perform the corresponding search.
Running these examples may change option settings.
- Legend – node-type color key,
with checkboxes to hide and show each type in the current network
Clicking Submit initiates the search, which may take several seconds
depending on the specific query and option settings.
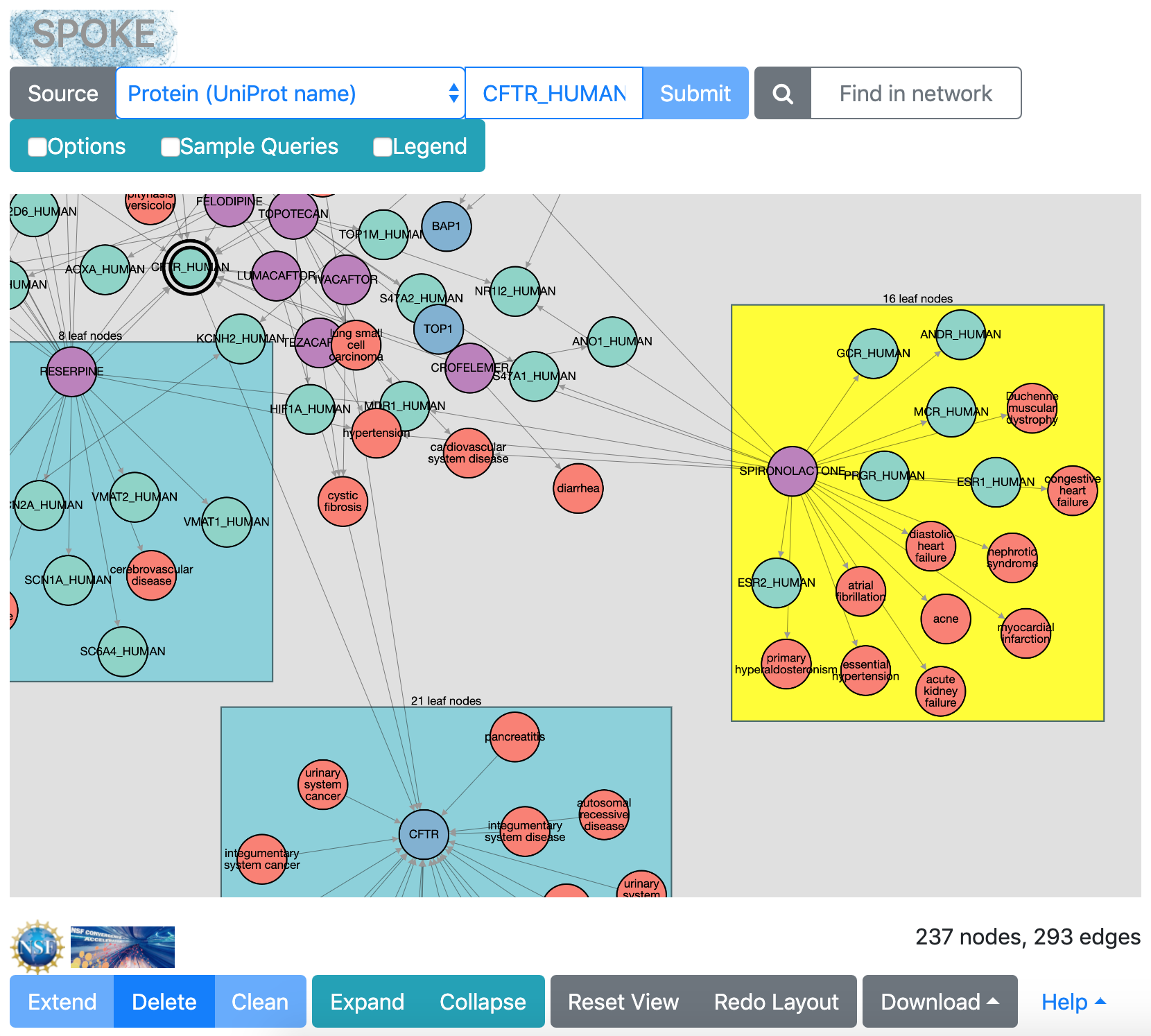
|
leaf-node group selected
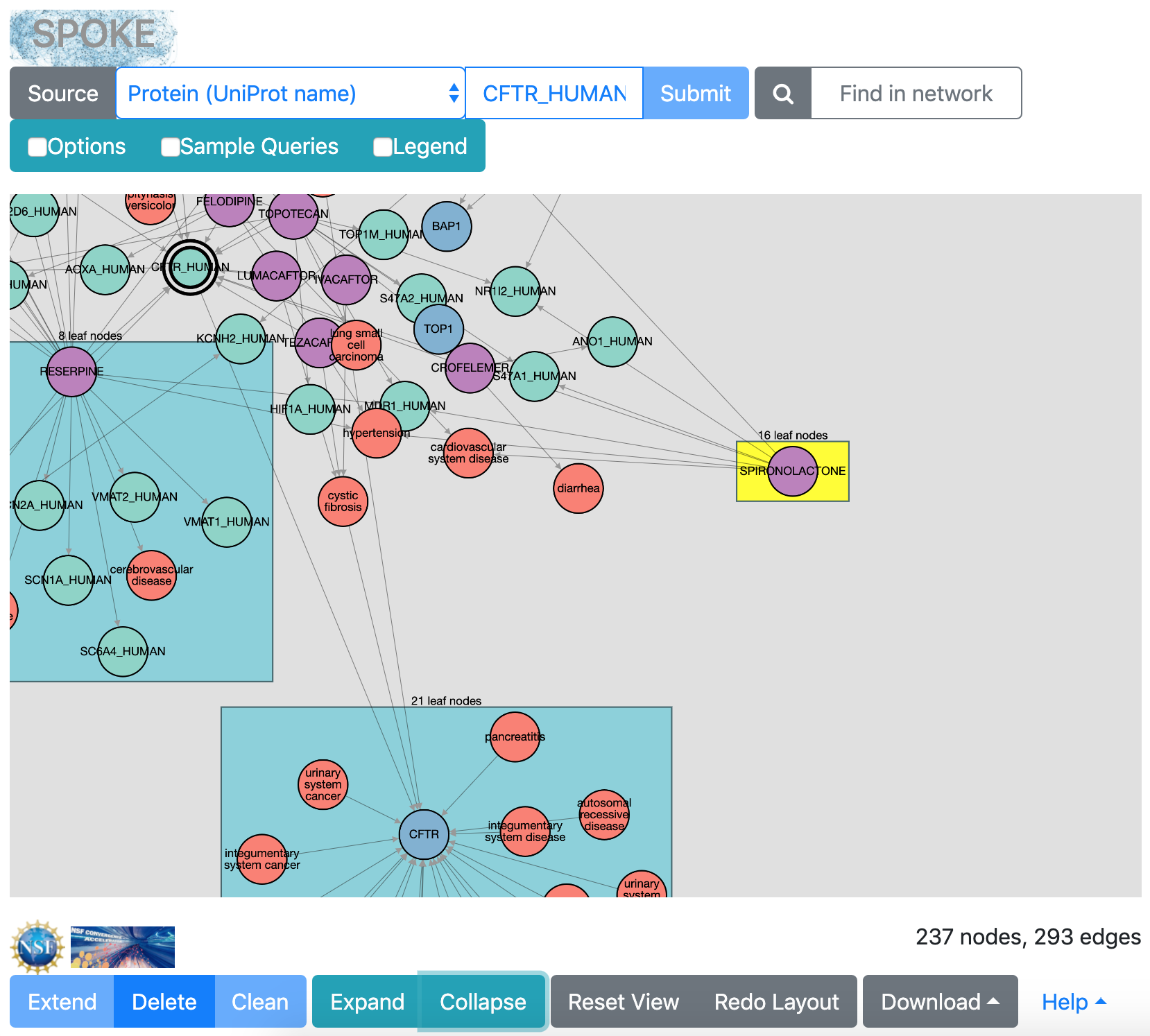
|
leaf-node group collapsed
|
 |
 |
[back to top]
Exploring the Network
The resulting network is displayed when the search is complete.
The query node is shown with a double border. Most edges are solid lines,
except that SEA search compound-protein binding predictions
are shown as dashed lines that vary in width by significance value.
General interactions with the network:
- Hovering the cursor over a node or edge shows its detailed information:
name, data source, etc.
- A node can be dragged to a different position in the network,
whereas starting the drag on “empty” space moves the entire network.
- Scrolling zooms the network.
- Clicking a node selects
it for subsequent button actions. Selected nodes are highlighted in yellow.
Shift-clicking a node toggles selection status or adds it to a pre-existing
selection.
- Another way to select nodes is by using the
“Find in network” text search box.
All nodes with the text string in their name or description will be selected.
- Clicking in empty space clears the selection.
In addition to the interactions mentioned above, the Neighborhood Explorer
provides several keyboard accelerators to manipulate node and edge selection,
move nodes around, and delete nodes:
| Key Combination | Action |
|---|
| Del |
Deletes the currently selected nodes |
↑ |
Moves selected nodes up |
Shift-↑ |
Moves selected nodes up a smaller increment |
↓ |
Moves selected nodes down |
Shift-↓ |
Moves selected nodes down a smaller increment |
← |
Moves selected nodes left |
Shift-← |
Moves selected nodes left a smaller increment |
→ |
Moves selected nodes right |
Shift-→ |
Moves selected nodes right a smaller increment |
Control-6 |
Selects the first neighbors of the selected nodes |
Control-I |
Inverts the current node selection |
Alt-I |
Inverts the current edge selection |
Alt-N |
Select All Nodes |
Alt-Shift-N |
Deselect All Nodes |
Alt-Control-N |
Selected nodes connected by selected edges |
Alt-Control-N |
Selected nodes connected by selected edges |
Alt-E |
Select All Edges |
Alt-Shift-E |
Deselect All Edges |
Alt-Control-E |
Select edges adjacent to selected nodes |
|
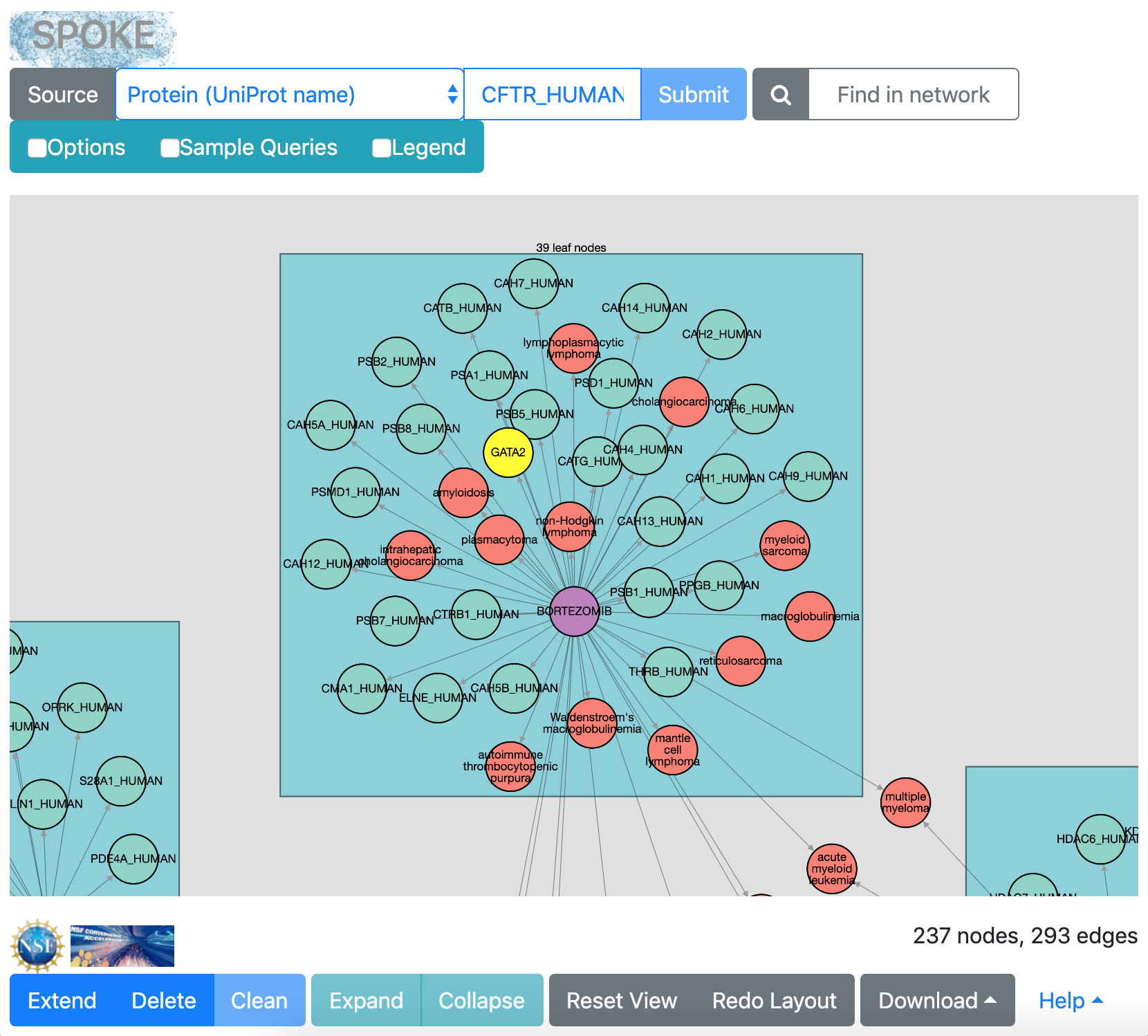
GATA2 gene node selected
|
after extending from GATA2 and redoing layout
|
 |
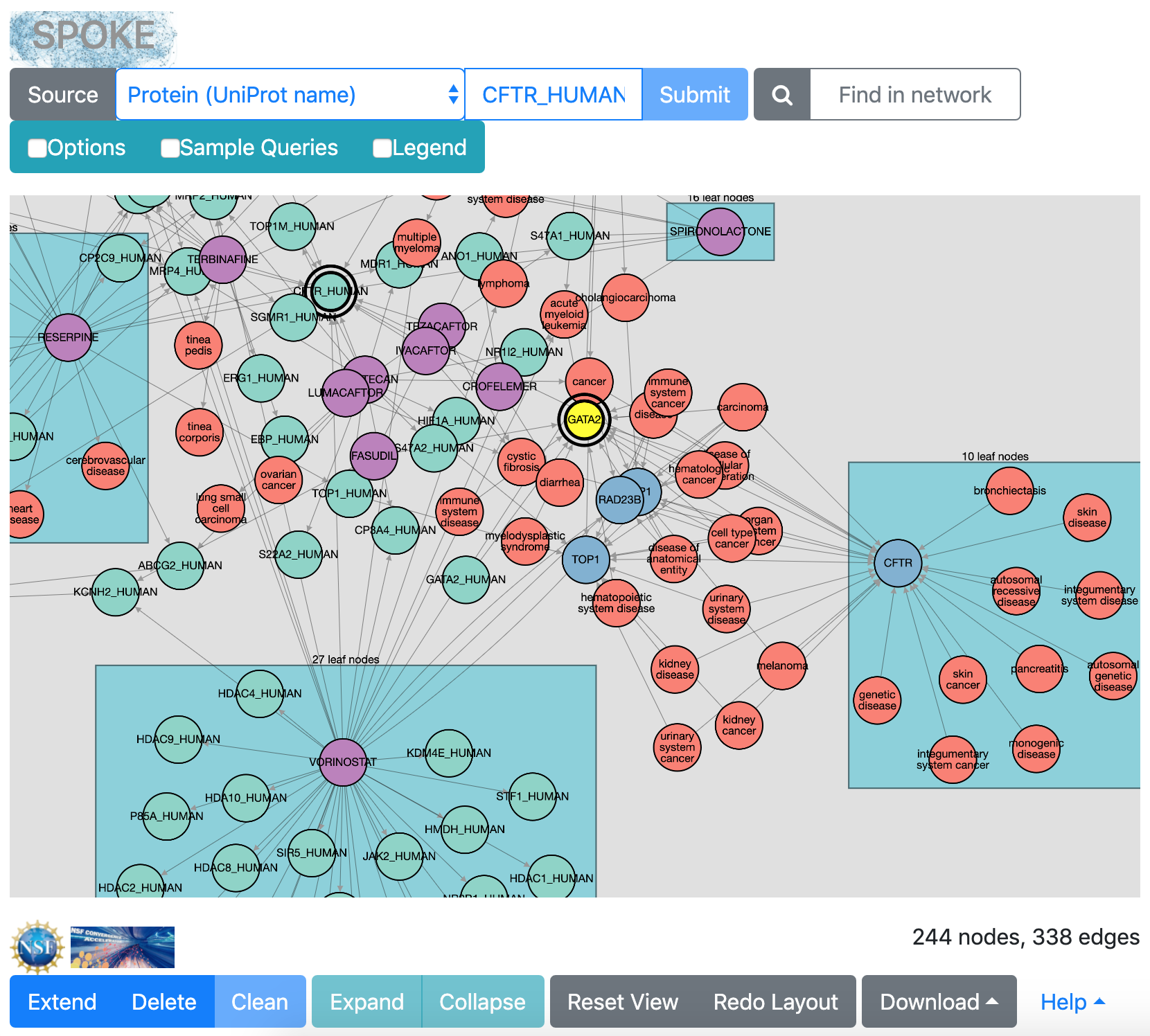 |
When numerous “leaf nodes” (those connected by only one edge)
emanate from the same central node, they are grouped into a rectangle
and can be collapsed onto that node to simplify the view.
Clicking the rectangle representing a leaf-node group
selects it for action by the Collapse button (or double-clicking),
or if the rectangle is in the collapsed state, for re-expansion with the
Expand button.
Other buttons:
- Extend the network by searching with the
selected node as query, using the current
option settings. Only one node should be selected.
- Delete selected nodes
- Clean – delete any nodes that cannot be reached through edges
from the double-border node(s)
- Reset View
– rescale and center the current network without changing the layout
- Redo Layout
– recalculate network layout
- Download
– download the network as files for Cytoscape (JSON and style) or an image (SVG or PNG)
- Help
– click to list choices:
- Documentation – open this documentation page
- Feedback – open a form to send comments or bug reports
on the Neighborhood Explorer
- SPOKE Home – show
SPOKE homepage
- SPOKE Version – show SPOKE update log
UCSF Resource for Biocomputing, Visualization, and Informatics /
October 2021






